Research Projects
Single-Molecule Orientation Localization Microscopy
Traditional fluorescence imaging measures the spatial structure of targets by focusing on measuring where lights come from.
In my projects, I measure both where lights come from and the shape of lights. This measurement gives us information on how fluorescent emitters interact with imaging targets, e.g. the binding direction (3D orientation) of fluorescence probes to target structure.
Point spread function engineering
Tingting Wu, Jin Lu, and Matthew D. Lew. Optica (2022). [Article] [Data&Code] [Summary video]
In this work, we “ruin” the microscope to generate point spread functions (PSFs) that deviate from the traditional Gaussian pattern. We designed an optimization algorithm to optimize a phase mask that we will put into the back focal plane of a polarized 4f microscope. The optimized microscope output PSFs with shapes vary distinctively for fluorescent probes with different orientations and axial positions.
The optimized microscope enables us to simultaneously measure the 3D position and 3D orientation of fluorescent probes. We reconstructed the shape of a spherical supported lipid bilayer (SLB). All the fluorescent emitters have orientation perpendicular to the spherical surface for SLB with cholesterol, however, once we delete the cholesterol, the SLB becomes more spacious for fluorescent probes to rotate. Therefore, we notice more random orientations for SLB without cholesterol.
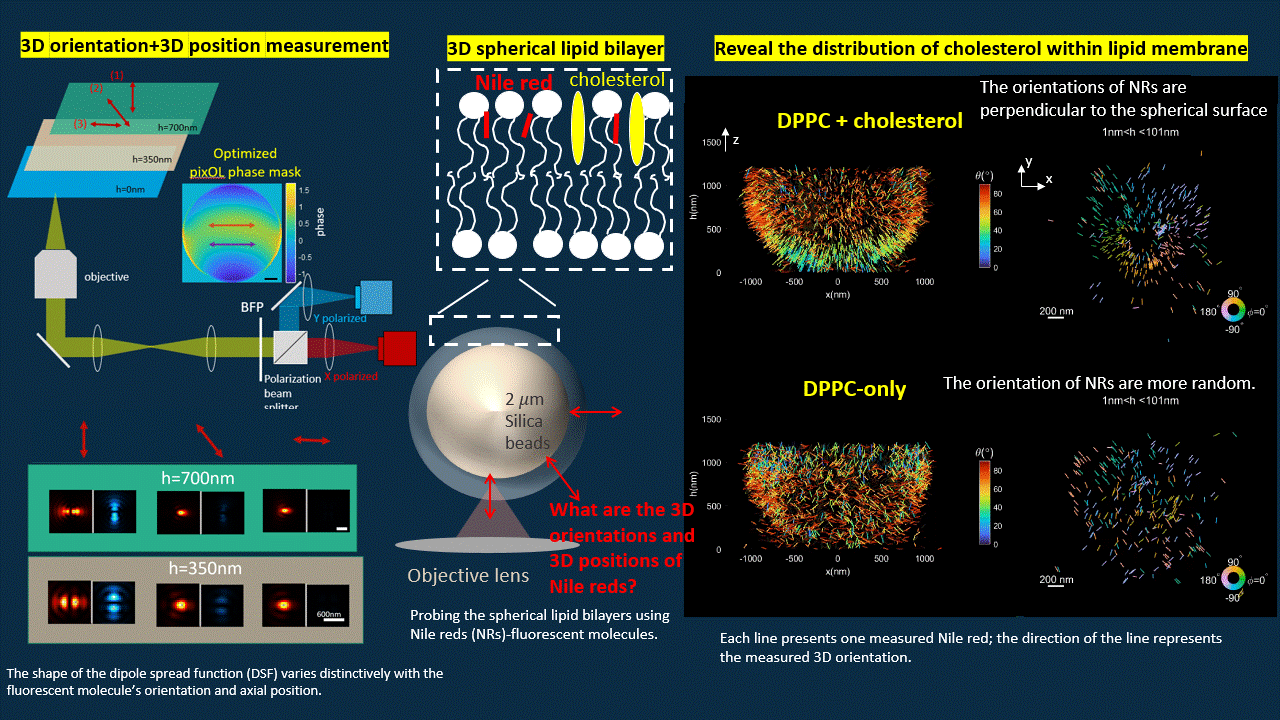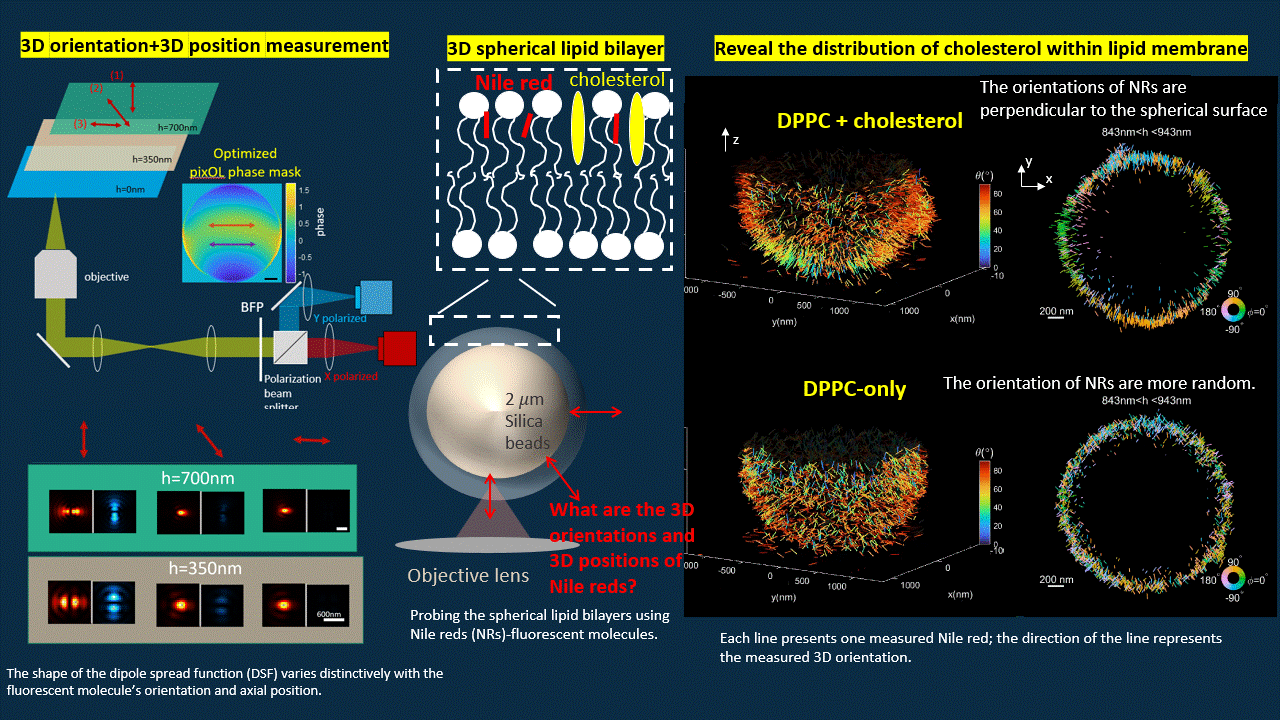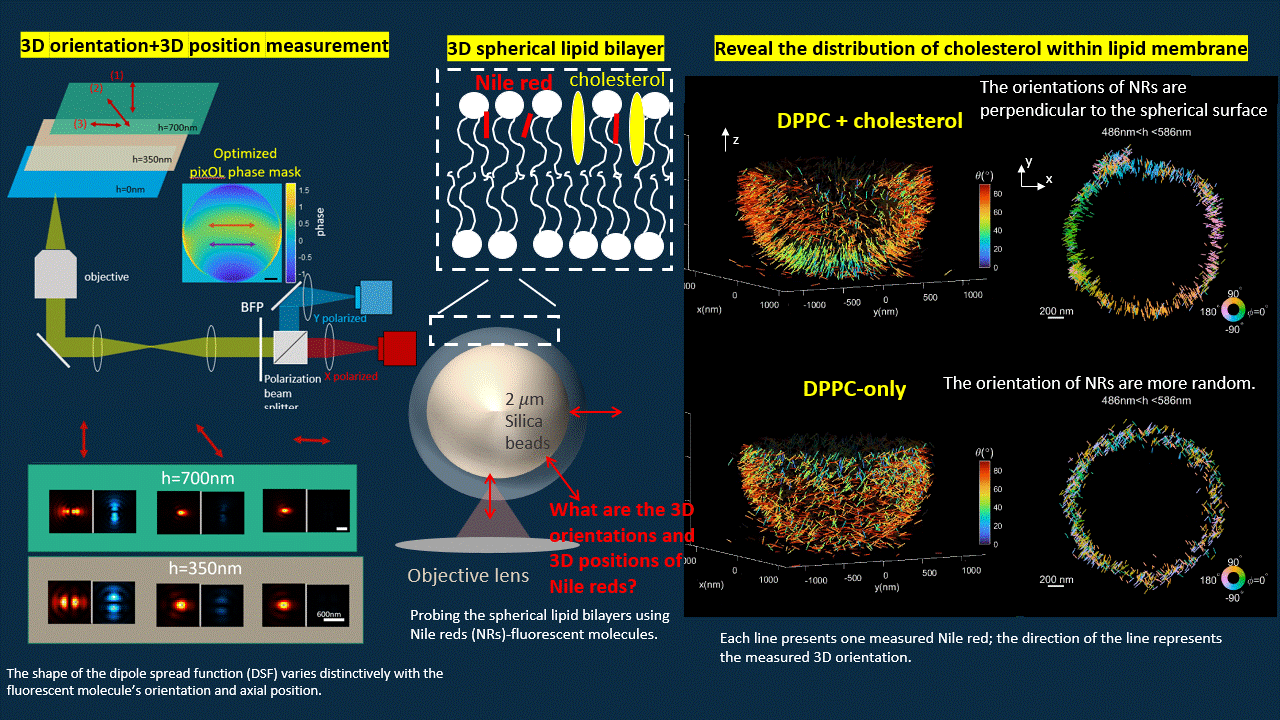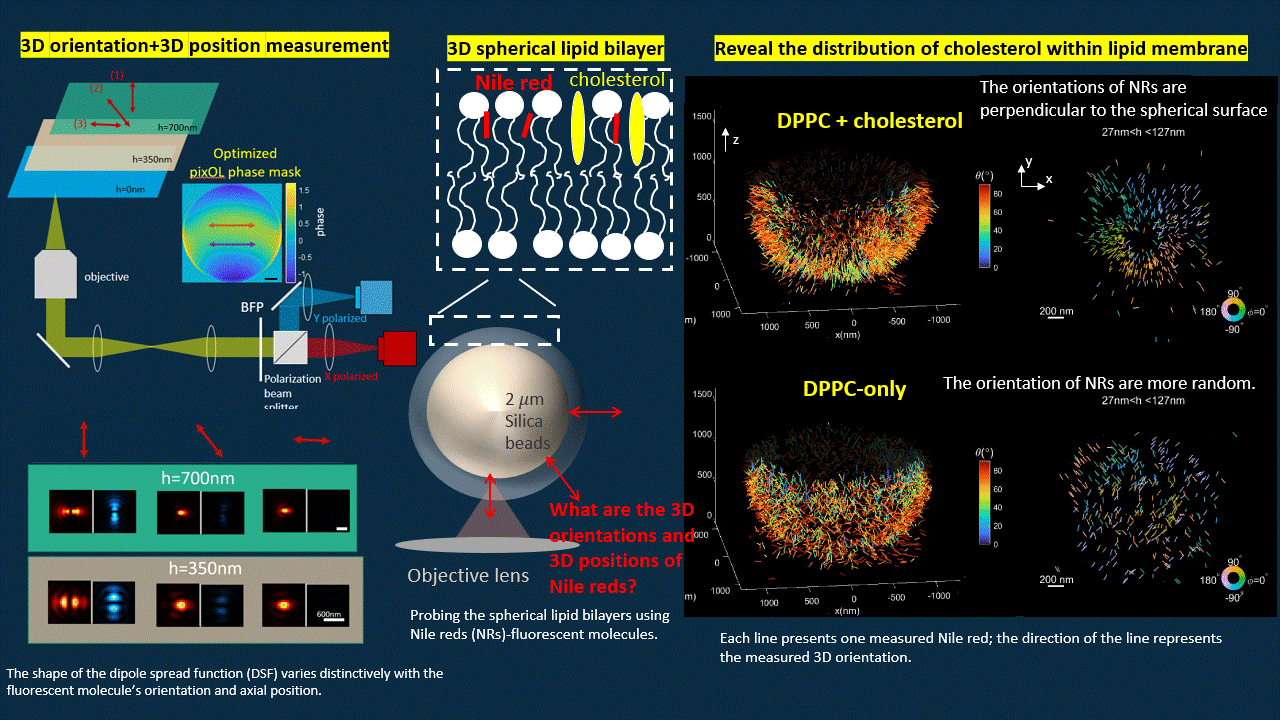
Deep-learning based estimation algorithm design
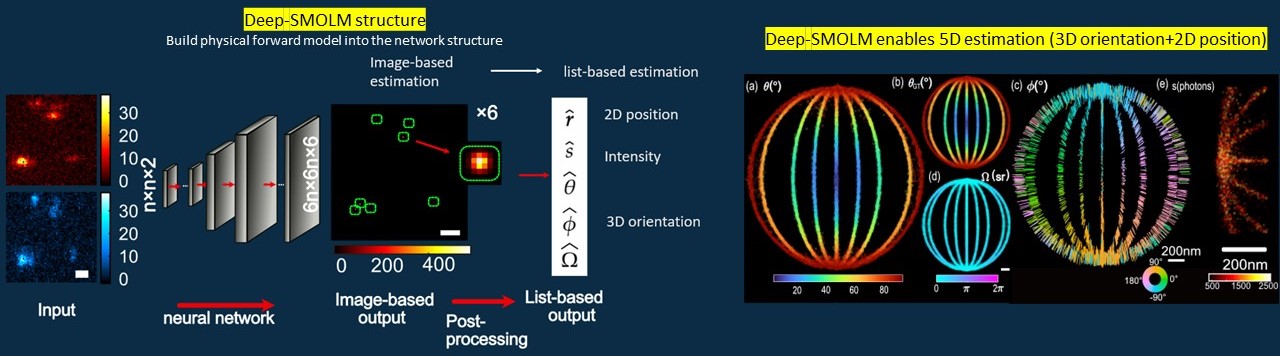
TingtingWu, Peng Lu†, Md Ashequr Rahman†, Xiao Li†, and Matthew D. Lew. Optics Express, in print (2022). [Article] [Code] [Data] [Summary video]
Using engineered PSF, emitters with different orientations will generate PSFs with different shapes on our camera. In this work, we design a deep-learning-based algorithm to estimate the 3D orientation and 2D position from the noise-crrupted PSFs.
We smartly designed our network to handle this high dimensional estimation challenge: 1) we encode the 3D orientation and 2D position orthogonally into the spatial position and intensity of the gaussian patterns on the output images; 2) we leverage the forward model to output the orientational second moments instead of the three orientation angles.
Deep-SMOLM outwins the iterative estimation algorithm in terms of 1) estimating overlapped emitters, 2) outputting precisions close to optimal due to its ability to give global minima instead of traping in the local minima, and 3) ~10 times faster estimation speed.
Adaptive microscopy design
ongoing project, more details will be added later
Most of the microsocpes are fixed once built. In this project, we configure the strucuture of the microscope to adapt to the current measurement to achieve better imaging.
Mapping heterogeneities inside biomolecule condensates using single molecule imaging and fluorogenic probes
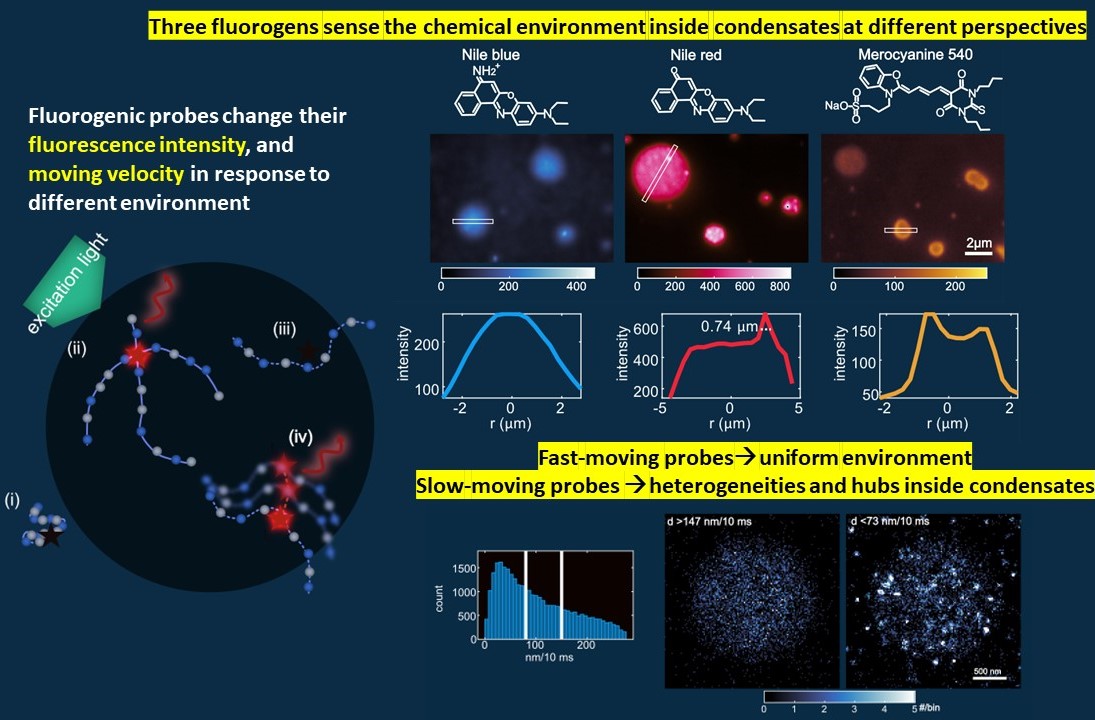
T. Wu, M.R. King, M. Farag, R. V. Pappue, and M. D. Lew, BioRxiv (2023). [Article].
Biomolecular condensates provide spatial and temporal control over biochemical reactions. Due to the high dynamic feature of condensates both spatially and temporally, the structures inside condensates haven’t been well explored
In this project, we deployed Nile blue (NB), Nile red (NR), and merocyanine 540 (MC540) for epifluorescence and single-molecule localization microscopy imaging of condensates formed by intrinsically disordered, low-complexity domains of proteins. Imaging with NB reveals internal environments that are uniformly hydrophobic, whereas NR shows preferential binding to hubs that are more hydrophobic than the surrounding background within condensates. Finally, imaging with MC540 suggests that interfaces of condensates are unique chemical environments. Overall, the high spatiotemporal resolution and environmental sensitivity of single-fluorogen imaging reveals spatially inhomogeneous organization of molecules within condensates.
Evaluation metric design based on information theory
Tianben Ding†, Tingting Wu†, Hesam Mazidi, Oumeng Zhang, and Matthew Lew. Optica 7.6 (2020) [Article] [Data&Code] [Summary slide]
In this project, we developed a metric, termed variance upper bound (VUB), to quickly select the best microscope for single-molecule orientation measurements. VUB, the global upper bound of Cramér-Rao bound (CRB) for all possible molecular orientations, efficiently quantifies the orientation sensitivity of a point spread function (PSF) ~1000X faster than calculating the average CRB over orientation space.
VUB enables us to realize that the polarized standard PSF (polar) provides superior measurement precision when molecules are near a refractive index (RI) interface. We then used the polarized standard PSF to measure the 3D orientation and 2D position of Nile red that transiently bind to the amyloid fibril.
Orientations of indivual Nile reds on amyloid fibril
Image credit to Prof. Matthew D. Lew and Dr. Tianben Ding
Optical fiber sensor
Tingting Wu, Linlin Xu, and Xinhai Zhang. Journal of Physics Communications 2.6 (2018), p. 065009. [Article].
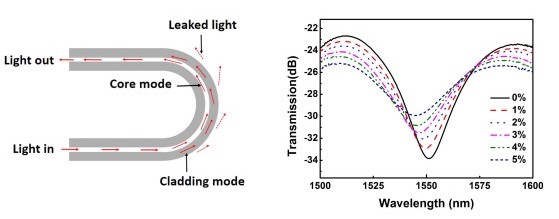 In this project, we designed a refractive index sensor by creating U-shaped optical fibers. The interference between the lights in the cladding mode and core mode, the optical fiber show different interference valley for solution with different refractive indexes.
In this project, we designed a refractive index sensor by creating U-shaped optical fibers. The interference between the lights in the cladding mode and core mode, the optical fiber show different interference valley for solution with different refractive indexes.
